Computes the exact Euler loglikelihood for any amount of missing data using a Kalman filter.
mou.loglik(X, dt, nvar.obs, Gamma, Lambda, Phi, mu0, Sigma0)
Arguments
| X | An |
|---|---|
| dt | A scalar or length |
| nvar.obs | A scalar or length |
| Gamma | A |
| Lambda | A length- |
| Phi | A |
| mu0, Sigma0 | Mean and variance of marginal multivariate normal distribution of |
Value
Scalar value of the loglikelihood. See Details.
Details
The \(p\)-dimensional multivariate Ornstein-Uhlenbeck (mOU) process \(Y_t = (Y_{1t}, \ldots, Y_{dt})\) satisfies the SDE
$$
dY_t = (\Gamma Y_t + \Lambda)dt + \Phi^{1/2} dB_t,
$$
where \(B_t = (B_{1t}, \ldots, B_{pt})\) is \(p\)-dimensional Brownian motion. Its Euler discretization is of the form
$$
Y_{n+1} = Y_n + (\Gamma Y_n + \Lambda) \Delta_n + \Phi^{1/2} \Delta B_n,
$$
where \(Y_n = Y(t_n)\), \(\Delta_n = t_{n+1} - t_n\) and
$$
\Delta B_n = B(t_{n+1}) - B(t_n) \stackrel{\textnormal{ind}}{\sim} \mathcal N(0, \Delta_n).
$$
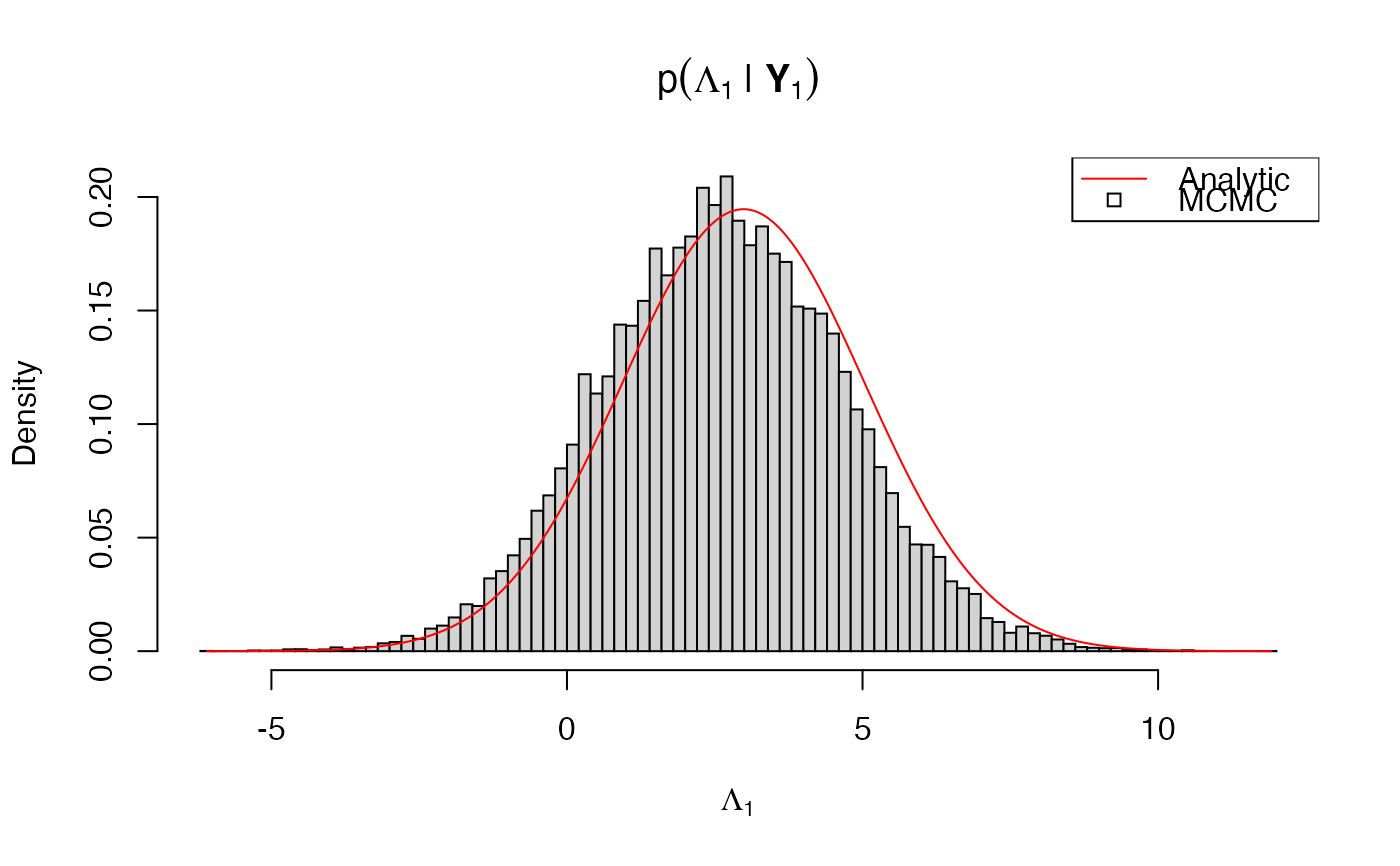
Thus, \(Y_0, \ldots, Y_N\) is multivariate normal Markov chain for which the marginal distribution of any subset of timepoints and/or components can be efficiently calculated using the Kalman filter. This can be used to check the MCMC output of sde.post() as in the example.
Examples
# \donttest{ # bivariate OU model bmod <- sde.examples("biou") # simulate some data # true parameter values Gamma0 <- .1 * crossprod(matrix(rnorm(4),2,2)) Lambda0 <- rnorm(2) Phi0 <- crossprod(matrix(rnorm(4),2,2)) Psi0 <- chol(Phi0) # precompiled model uses the Cholesky scale theta0 <- c(Gamma0, Lambda0, Psi0[c(1,3,4)]) names(theta0) <- bmod$param.names # initial value Y0 <- rnorm(2) names(Y0) <- bmod$data.names # simulation dT <- runif(1, max = .1) # time step nObs <- 10 bsim <- sde.sim(bmod, x0 = Y0, theta = theta0, dt = dT, dt.sim = dT, nobs = nObs)#>#>#>#>YObs <- bsim$data # inference via MCMC binit <- sde.init(bmod, x = YObs, dt = dT, theta = theta0, nvar.obs = 1) # second component is unobserved # only Lambda1 is unknown fixed.params <- rep(TRUE, bmod$nparams) names(fixed.params) <- bmod$param.names fixed.params["Lambda1"] <- FALSE # prior on (Lambda1, Y_0) hyper <- list(mu = c(0,0), Sigma = diag(2)) names(hyper$mu) <- bmod$data.names dimnames(hyper$Sigma) <- rep(list(bmod$data.names), 2) # posterior sampling nsamples <- 1e5 burn <- 1e3 bpost <- sde.post(bmod, binit, hyper = hyper, fixed.params = fixed.params, nsamples = nsamples, burn = burn)#>#>#>#>#>#>#>#>L1.mcmc <- bpost$params[,"Lambda1"] # analytic posterior L1.seq <- seq(min(L1.mcmc), max(L1.mcmc), len = 500) L1.loglik <- sapply(L1.seq, function(l1) { lambda <- Lambda0 lambda[1] <- l1 mou.loglik(X = YObs, dt = dT, nvar.obs = 1, Gamma = Gamma0, Lambda = lambda, Phi = Phi0, mu0 = hyper$mu, Sigma0 = hyper$Sigma) }) # normalize density L1.Kalman <- exp(L1.loglik - max(L1.loglik)) L1.Kalman <- L1.Kalman/sum(L1.Kalman)/(L1.seq[2]-L1.seq[1]) # compare hist(L1.mcmc, breaks = 100, freq = FALSE, main = expression(p(Lambda[1]*" | "*bold(Y)[1])), xlab = expression(Lambda[1]))legend("topright", legend = c("Analytic", "MCMC"), pch = c(NA, 22), lty = c(1, NA), col = c("red", "black"))# }