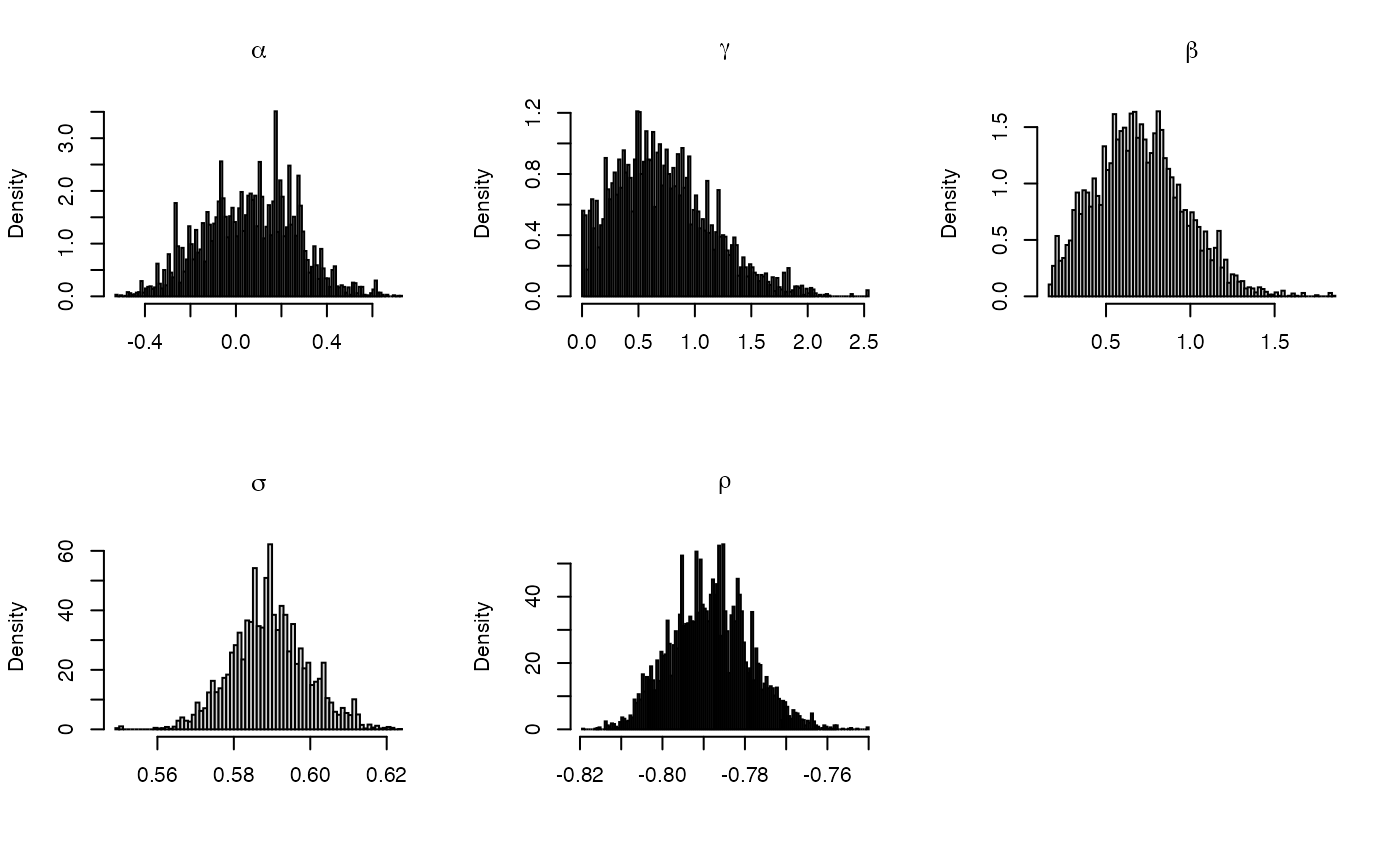A Metropolis-within-Gibbs sampler for the Euler-Maruyama approximation to the true posterior density.
sde.post( model, init, hyper, nsamples, burn, mwg.sd = NULL, adapt = TRUE, loglik.out = FALSE, last.miss.out = FALSE, update.data = TRUE, data.out, update.params = TRUE, fixed.params, ncores = 1, verbose = TRUE )
Arguments
| model | An |
|---|---|
| init | An |
| hyper | The hyperparameters of the SDE prior. See |
| nsamples | Number of MCMC iterations. |
| burn | Integer number of burn-in samples, or fraction of |
| mwg.sd | Standard deviation jump size for Metropolis-within-Gibbs on parameters and missing components of first SDE observation (see Details). |
| adapt | Logical or list to specify adaptive Metropolis-within-Gibbs sampling (see Details). |
| loglik.out | Logical, whether to return the loglikelihood at each step. |
| last.miss.out | Logical, whether to return the missing sde components of the last observation. |
| update.data | Logical, whether to update the missing data. |
| data.out | A scalar, integer vector, or list of three integer vectors determining the subset of data to be returned (see Details). |
| update.params | Logical, whether to update the model parameters. |
| fixed.params | Logical vector of length |
| ncores | If |
| verbose | Logical, whether to periodically output MCMC status. |
Value
A list with elements:
paramsAn
nsamples x nparamsmatrix of posterior parameter draws.dataA 3-d array of posterior missing data draws, for which the output dimensions are specified by
data.out.initThe
sde.initobject which initialized the sampler.data.outA list of three integer vectors specifying which timepoints, variables, and MCMC iterations correspond to the values in the
dataoutput.mwg.sdA named vector of Metropolis-within-Gibbs standard devations used at the last posterior iteration.
hyperThe hyperparameter specification.
loglikIf
loglik.out == TRUE, the vector ofnsamplescomplete data loglikelihoods calculated at each posterior sample.last.iterA list with elements
dataandparamsgiving the last MCMC sample. Useful for resuming the MCMC from that point.last.missIf
last.miss.out == TRUE, annsamples x nmissNmatrix of all posterior draws for the missing data in the final observation. Useful for SDE forecasting at future timepoints.acceptA named list of acceptance rates for the various components of the MCMC sampler.
Details
The Metropolis-within-Gibbs (MWG) jump sizes can be specified as a scalar, a vector or length nparams + ndims, or a named vector containing the elements defined by sde.init$nvar.obs.m[1] (the missing variables in the first SDE observation) and fixed.params (the SDE parameters which are not held fixed). The default jump sizes for each MWG random variable are .25 * |initial_value| when |initial_value| > 0, and 1 otherwise.
adapt == TRUE implements an adaptive MCMC proposal by Roberts and Rosenthal (2009). At step \(n\) of the MCMC, the jump size of each MWG random variable is increased or decreased by \(\delta(n)\), depending on whether the cumulative acceptance rate is above or below the optimal value of 0.44. If \(\sigma_n\) is the size of the jump at step \(n\), then the next jump size is determined by
$$
\log(\sigma_{n+1}) = \log(\sigma_n) \pm \delta(n), \qquad \delta(n) = \min(.01, 1/n^{1/2}).
$$
When adapt is not logical, it is a list with elements max and rate, such that delta(n) = min(max, 1/n^rate). These elements can be scalars or vectors in the same manner as mwg.sd.
For SDE models with thousands of latent variables, data.out can be used to thin the MCMC missing data output. An integer vector or scalar returns specific or evenly-spaced posterior samples from the ncomp x ndims complete data matrix. A list with elements isamples, icomp, and idims determines which samples, time points, and SDE variables to return. The first of these can be a scalar or vector with the same meaning as before.
References
Roberts, G.O. and Rosenthal, J.S. "Examples of adaptive MCMC." Journal of Computational and Graphical Statistics 18.2 (2009): 349-367. http://www.probability.ca/jeff/ftpdir/adaptex.pdf.
Examples
# \donttest{ # Posterior inference for Heston's model hmod <- sde.examples("hest") # load pre-compiled model # Simulate data X0 <- c(X = log(1000), Z = 0.1) theta <- c(alpha = 0.1, gamma = 1, beta = 0.8, sigma = 0.6, rho = -0.8) dT <- 1/252 nobs <- 1000 hest.sim <- sde.sim(model = hmod, x0 = X0, theta = theta, dt = dT, dt.sim = dT/10, nobs = nobs)#>#>#>#># initialize MCMC sampler # both components observed, no missing data between observations init <- sde.init(model = hmod, x = hest.sim$data, dt = hest.sim$dt, theta = theta) # Initialize posterior sampling argument nsamples <- 1e4 burn <- 1e3 hyper <- NULL # flat prior hest.post <- sde.post(model = hmod, init = init, hyper = hyper, nsamples = nsamples, burn = burn)#>#>#>#>#>#>#>#>#># plot the histogram for the sampled parameters par(mfrow = c(2,3)) for(ii in 1:length(hmod$param.names)) { hist(hest.post$params[,ii],breaks=100, freq = FALSE, main = parse(text = hmod$param.names[ii]), xlab = "") } # }