Estimate the bivariate copula dependence parameter eta at multiple covariate values.
CondiCopLocFit(
u1,
u2,
family,
x,
x0,
nx = 100,
degree = 1,
eta,
nu,
kernel = KernEpa,
band,
optim_fun,
cl = NA
)Arguments
- u1
Vector of first uniform response.
- u2
Vector of second uniform response.
- family
An integer defining the bivariate copula family to use. See
ConvertPar().- x
Vector of observed covariate values.
- x0
Vector of covariate values within
range(x)at which to fit the local likelihood. Does not have to be a subset ofx.- nx
If
x0is missing, defaults tonxequally spaced values inrange(x).- degree
Integer specifying the polynomial order of the local likelihood function. Currently only 0 and 1 are supported.
- eta
Optional initial value of the copula dependence parameter (scalar). If missing will be estimated unconditionally by
VineCopula::BiCopEst().- nu
Optional initial value of second copula parameter, if it exists. If missing and required, will be estimated unconditionally by
VineCopula::BiCopEst(). If provided and required, will not be estimated.- kernel
Kernel function to use. Should accept a numeric vector parameter and return a non-negative numeric vector of the same length. See
KernFun().- band
Kernal bandwidth parameter (positive scalar). See
KernWeight().- optim_fun
Optional specification of local likelihood optimization algorithm. See Details.
- cl
Optional parallel cluster created with
parallel::makeCluster(), in which case optimization for each element ofx0will be done in parallel on separate cores. Ifcl == NA, computations are run serially.
Value
List with the following elements:
xThe vector of covariate values
x0at which the local likelihood is fit.etaThe vector of estimated dependence parameters of the same length as
x0.nuThe scalar value of the estimated (or provided) second copula parameter.
Details
By default, optimization is performed with the quasi-Newton algorithm provided by stats::nlminb(), which uses gradient information provided by automatic differentiation (AD) as implemented by TMB.
If the default method is to be overridden, optim_fun should be provided as a function taking a single argument corresponding to the output of CondiCopLocFun(), and return a scalar value corresponding to the estimate of eta at a given covariate value in x0. Note that TMB calculates the negative local (log)likelihood, such that the objective function is to be minimized. See Examples.
Examples
# simulate data
family <- 5 # Frank copula
n <- 1000
x <- runif(n) # covariate values
eta_fun <- function(x) 2*cos(12*pi*x) # copula dependence parameter
eta_true <- eta_fun(x)
par_true <- BiCopEta2Par(family, eta = eta_true)
udata <- VineCopula::BiCopSim(n, family=family,
par = par_true$par)
# local likelihood estimation
x0 <- seq(min(x), max(x), len = 100)
band <- .02
system.time({
eta_hat <- CondiCopLocFit(u1 = udata[,1], u2 = udata[,2],
family = family, x = x, x0 = x0, band = band)
})
#> user system elapsed
#> 0.19 0.01 0.20
# custom optimization routine using stats::optim (gradient-free)
my_optim <- function(obj) {
opt <- stats::optim(par = obj$par, fn = obj$fn, method = "Nelder-Mead")
return(opt$par[1]) # always return constant term, even if degree > 0
}
system.time({
eta_hat2 <- CondiCopLocFit(u1 = udata[,1], u2 = udata[,2],
family = family, x = x, x0 = x0, band = band,
optim_fun = my_optim)
})
#> user system elapsed
#> 0.374 0.014 0.389
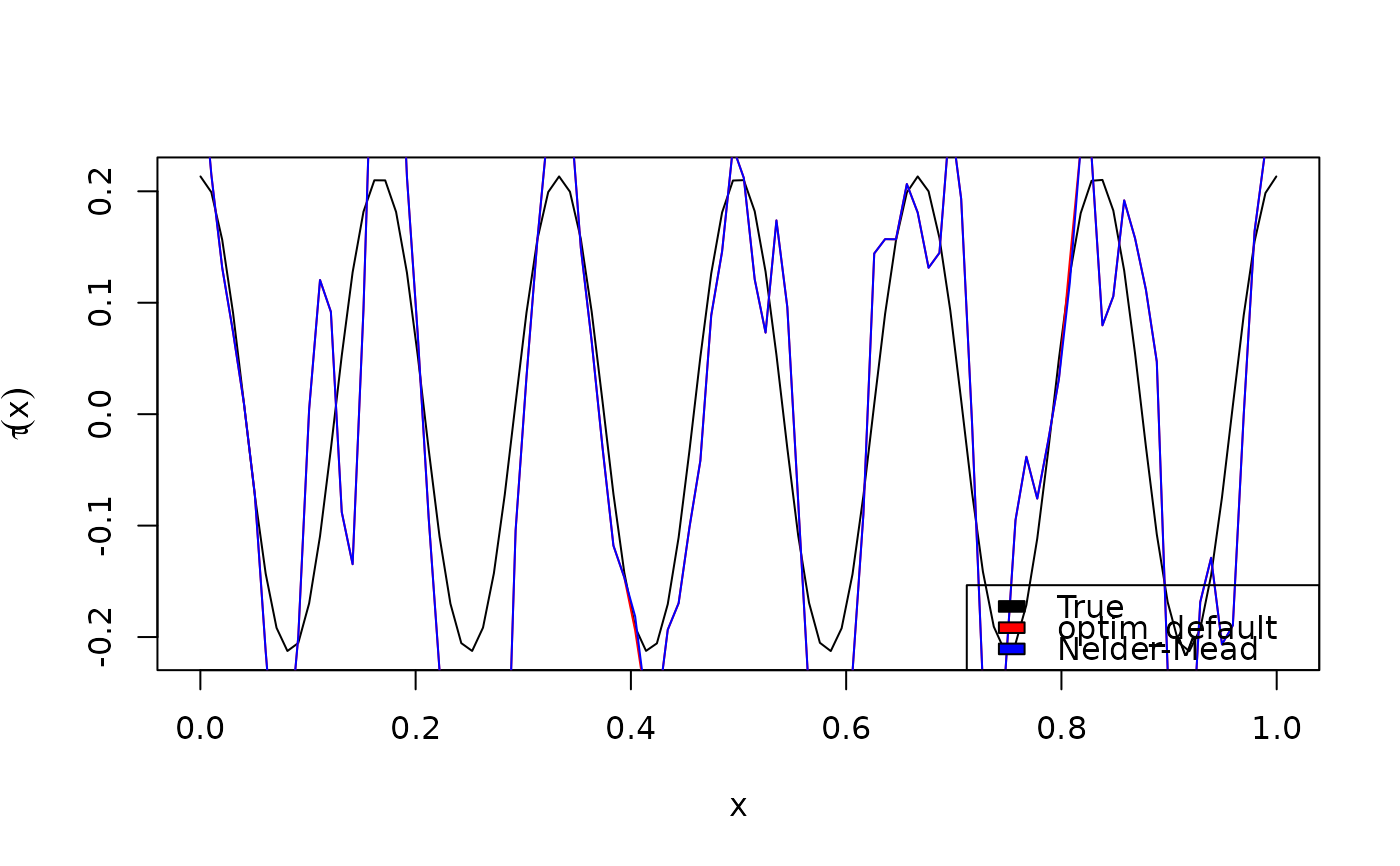
plot(x0, BiCopEta2Tau(family, eta = eta_fun(x0)), type = "l",
xlab = expression(x), ylab = expression(tau(x)))
lines(x0, BiCopEta2Tau(family, eta = eta_hat$eta), col = "red")
lines(x0, BiCopEta2Tau(family, eta = eta_hat2$eta), col = "blue")
legend("bottomright", fill = c("black", "red", "blue"),
legend = c("True", "optim_default", "Nelder-Mead"))