Local likelihood inference for conditional copula models.
Source:R/LocalCop-package.R
LocalCop-package.RdFits a bivariate conditional copula \(C(u_1, u_2 | \theta_x)\), where \(\theta_x\) is a variable dependence parameter, nonparametrically estimated from a single covariate \(x\) via local likelihood.
See also
Useful links:
Examples
# simulate data
set.seed(123)
family <- 5 # Frank copula
n <- 1000
x <- runif(n) # covariate values
eta_fun <- function(x) 2*cos(12*pi*x) # copula dependence parameter
eta_true <- eta_fun(x)
par_true <- BiCopEta2Par(family, eta = eta_true)
udata <- VineCopula::BiCopSim(n, family=family,
par = par_true$par)
# bandwidth and family selection
bandset <- c(.01, .04, .1) # bandwidth set
famset <- c(2, 5) # family set
n_loo <- 100 # number of leave-one-out observations in CV likelihood calculation
system.time({
cvsel <- CondiCopSelect(u1= udata[,1], u2 = udata[,2],
x = x, family = famset, band = bandset,
xind = n_loo)
})
#> user system elapsed
#> 1.766 0.004 1.770
# compare estimates to true value
xseq <- cvsel$x
famsel <- cvsel$cv$family
bandsel <- cvsel$cv$band
etasel <- cvsel$eta
clrs <- c("red", "blue", "green4")
names(clrs) <- bandset
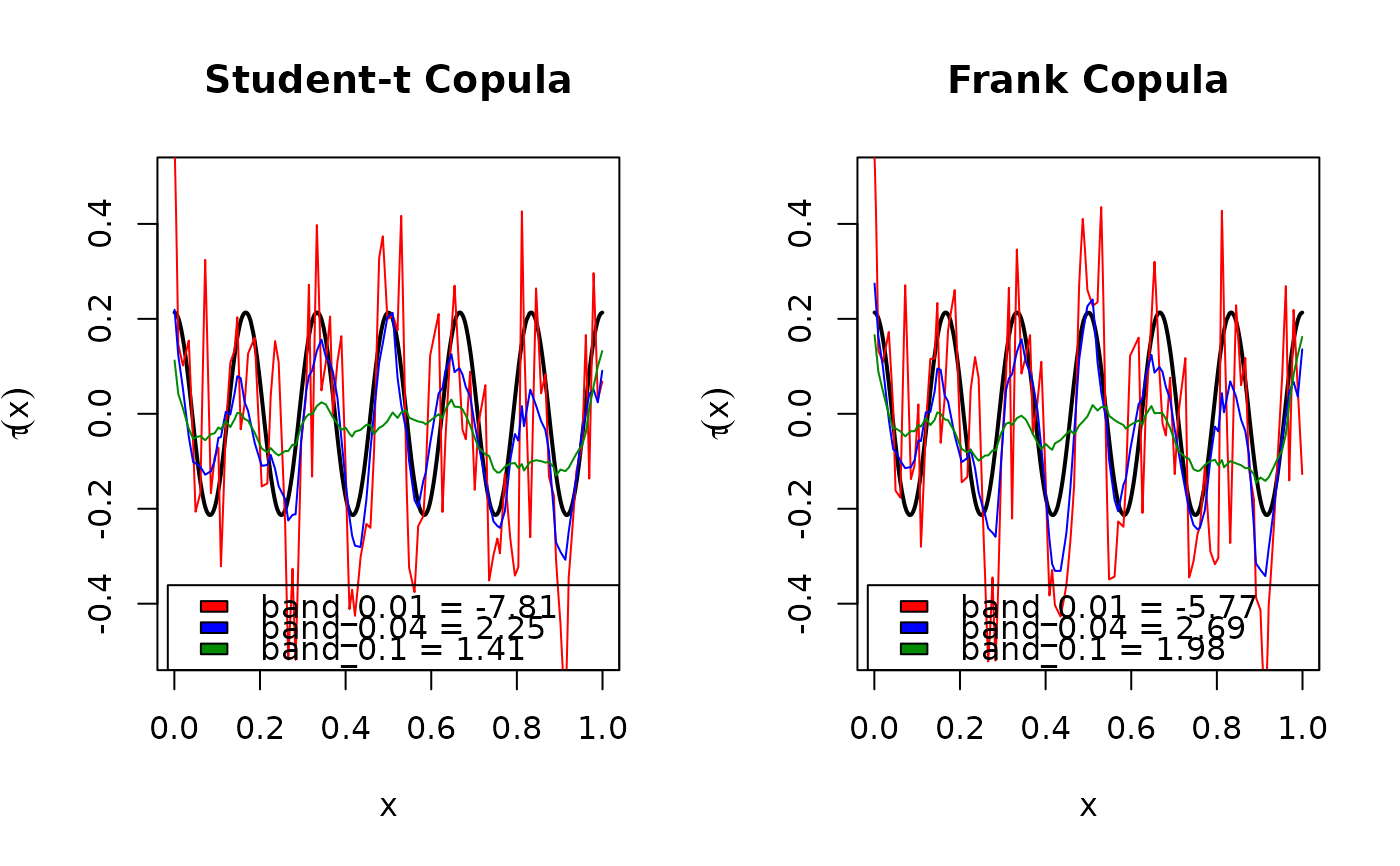
plot_fun <- function(fam) {
nband <- length(bandset)
if(fam == 2) {
famind <- 1:nband
main <- "Student-t Copula"
} else {
famind <- nband+1:nband
main <- "Frank Copula"
}
plot(xseq, BiCopEta2Tau(family, eta = eta_fun(xseq)),
type = "l", lwd = 2, ylim = c(-.5, .5),
xlab = expression(x), ylab = expression(tau(x)),
main = main)
for(ii in famind) {
lines(xseq, BiCopEta2Tau(fam, eta = etasel[,ii]),
col = clrs[as.character(bandsel[ii])], lwd = 1)
}
legend("bottomright", fill = clrs,
legend = paste0("band_", bandsel[famind],
" = ", signif(cvsel$cv$cv[famind], 3)))
}
oldpar <- par(mfrow = c(1,2))
plot_fun(2)
plot_fun(5)
 par(oldpar)
par(oldpar)